\(\newcommand{L}[1]{\| #1 \|}\newcommand{VL}[1]{\L{ \vec{#1} }}\newcommand{R}[1]{\operatorname{Re}\,(#1)}\newcommand{I}[1]{\operatorname{Im}\, (#1)}\)
Find the arteries solution¶
>>> #: Our usual imports
>>> import numpy as np # Python array library
>>> import matplotlib.pyplot as plt # Python plotting library
The major arteries in a T1 MRI image often have high signal (white when displaying in gray-scale).
Our task is to see if we can pick out the courses of the vertebral, basilar and carotid arteries on this image.
The image is ds107_sub001_highres.nii. Download
ds107_sub001_highres.nii to your working directory.
This time we are going to load the image using the nibabel library.
>>> #: Import nibabel
>>> import nibabel as nib
Try loading the image using nibabel, to make an image object. Use tab
completion on nib. in IPython to see if you can fund the function you
need. Go back to have a look at What is an image? if
you are stuck.
>>> #- Use nibabel to load the image ds107_sub001_highres.nii
>>> #- img = ?
>>> img = nib.load('ds107_sub001_highres.nii')
Now get the image array data from the nibabel image object. Don’t forget to use tab completion on the image object if you can’t remember or don’t know the methods of the object.
>>> #- data = ?
>>> data = img.get_data()
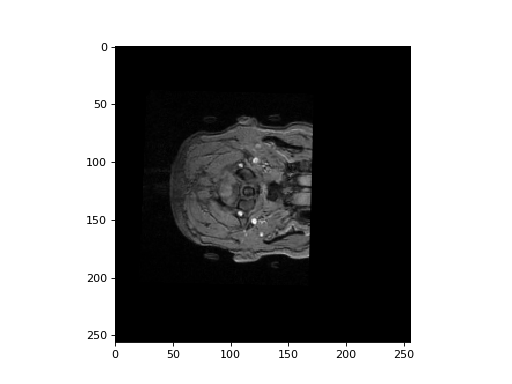
Try plotting a few slices over the third dimension to see whether you can see
the arteries. For example, if data is your image array data, then you
might plot the first slice over the third dimension, you might use:
plt.imshow(data[:, :, 0], cmap='gray')
>>> #- Plotting some slices over the third dimension
>>> plt.imshow(data[:, :, 30], cmap='gray')
<...>
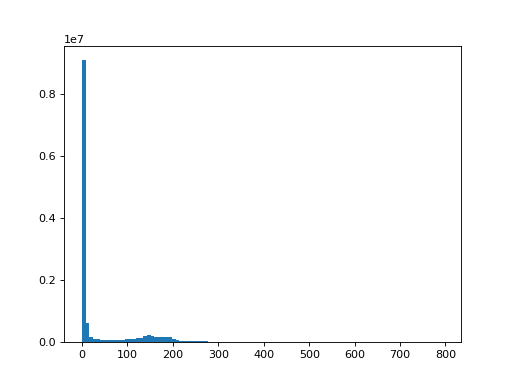
Now try looking for a good threshold so that you pick up only the very high
signal in the brain. A good place to start is to use plt.hist to get an
idea of the spread of values within the volume and within the slice.
>>> #- Here you might try plt.hist or something else to find a threshold
>>> data_1d = data.ravel()
>>> plt.hist(data_1d, bins=100)
(...)
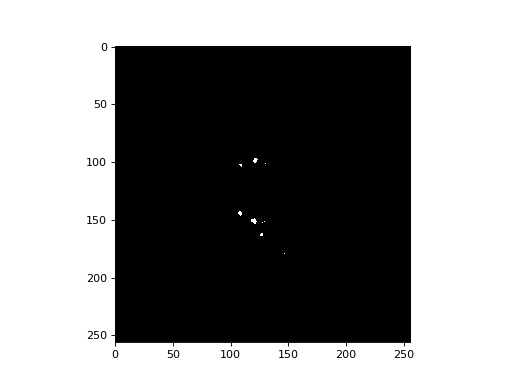
Try making a binarized image with your threshold and displaying slices with that. What structures are you picking up?
>>> #- Maybe display some slices from the data binarized with a threshold
>>> threshold = 300
>>> binarized_data = data > threshold
>>> plt.imshow(binarized_data[:, :, 30], cmap='gray')
<...>
Now try taking a 3D subvolume out of the middle of the image. Take the approximate middle in all three axes. Use this to pick out a good subvolume of the image that still contains the big arteries.
>>> #- Create a smaller 3D subvolume from the image data that still
>>> #- contains the arteries
>>> subvolume = data[90:-90, 90:-90, 10:130]
Try binarizing the subvolume with some thresholds to see whether you can pick
out the arteries without much other stuff. Hint - you might consider using
np.percentile or plt.hist to find a good threshold.
>>> #- Try a few plots of binarized slices and other stuff to find a good
>>> #- threshold
>>> pct_99 = np.percentile(subvolume, 99)
>>> binarized_subvolume = subvolume > pct_99
>>> plt.imshow(binarized_subvolume[:, :, 20], cmap='gray')
<...>
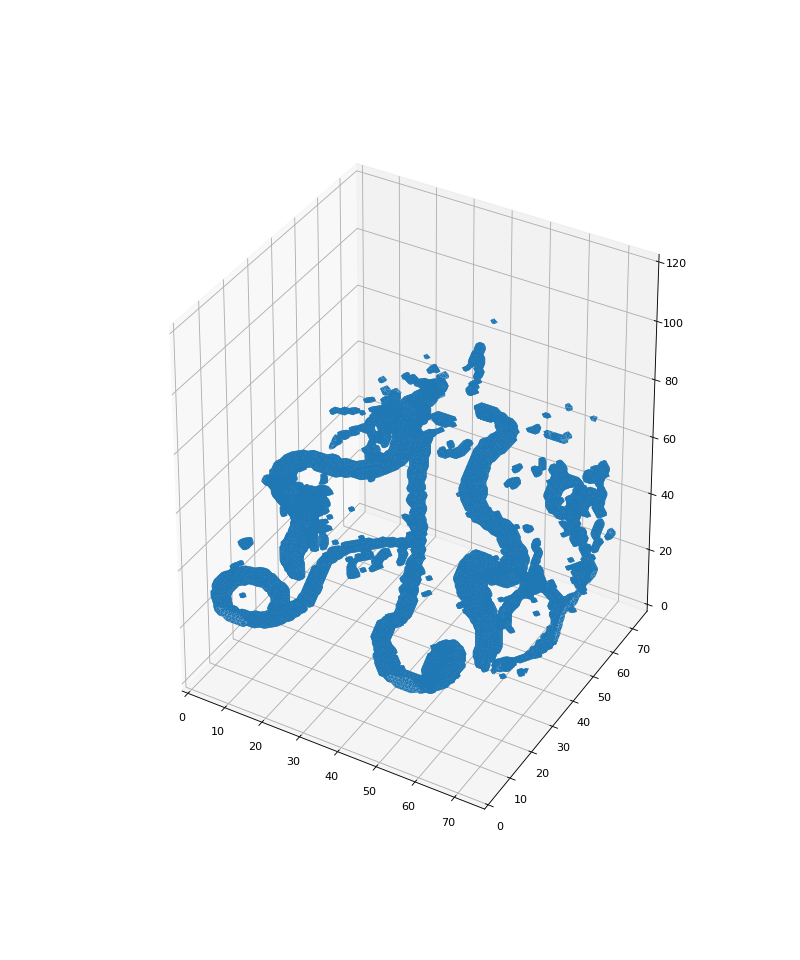
If you have a good threshold and a good binarized subset, see if you can see
the arterial structure using the following fancy function to plot the
binarized image with a 3D rendering. To use this function, you will need to
install the scikit-image toolbox. First see if scikit-image is installed
with the command import skimage from the Python / IPython console. If
this gives you an ImportError, then open a new terminal window and
install scikit-image with:
pip3 install --user scikit-image
Careful – do not run this pip3 install command from Python / IPython,
but from the terminal command window.
When you have done the scikit-image install, uncomment this code:
>>> #: Uncomment the next line after installing scikit-image
>>> # from skimage import measure
With that import done, here is the fancy function to display your subvolume in 3D:
>>> #: This function uses matplotlib 3D plotting and sckit-image for
>>> # rendering
>>> from mpl_toolkits.mplot3d.art3d import Poly3DCollection
>>>
>>> def binarized_surface(binary_array):
... """ Do a 3D plot of the surfaces in a binarized image
...
... The function does the plotting with scikit-image and some fancy
... commands that we don't need to worry about at the moment.
... """
... # Here we use the scikit-image "measure" function
... verts, faces, norms, vals = mcubes(binary_array, 0)
... fig = plt.figure(figsize=(10, 12))
... ax = fig.add_subplot(111, projection='3d')
...
... # Fancy indexing: `verts[faces]` to generate a collection of triangles
... mesh = Poly3DCollection(verts[faces], linewidths=0, alpha=0.5)
... ax.add_collection3d(mesh)
... ax.set_xlim(0, binary_array.shape[0])
... ax.set_ylim(0, binary_array.shape[1])
... ax.set_zlim(0, binary_array.shape[2])
For example, let’s say you have a binarized subvolume of the original
data called binarized_subvolume. You could do a 3D rendering of this
binary image with:
binarized_surface(binarized_subvolume)
>>> binarized_surface(binarized_subvolume)